LATEST RESEARCH
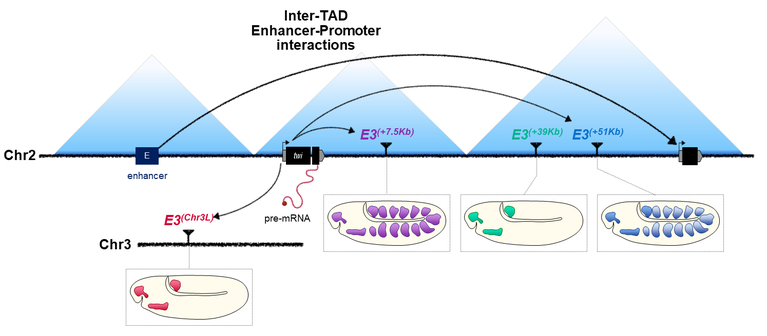
Enhancer–promoter interactions can form independently of genomic distance and be functional across TAD boundaries
Topologically Associating Domains (TADs) have been suggested to facilitate and constrain enhancer–promoter interactions. However, the role of TAD boundaries in effectively restricting these interactions remains unclear. Here, we show that a significant proportion of enhancer–promoter interactions are established across TAD boundaries in Drosophila embryos, but that developmental genes are strikingly enriched in intra- but not inter-TAD interactions. We pursued this observation using the twist locus, a master regulator of mesoderm development, and systematically relocated one of its enhancers to various genomic locations. While this developmental gene can establish inter-TAD interactions with its enhancer, the functionality of these interactions remains limited, highlighting the existence of topological constraints. Furthermore, contrary to intra-TAD interactions, the formation of inter-TAD enhancer–promoter interactions is not solely driven by genomic distance, with distal interactions sometimes favored over proximal ones. These observations suggest that other general mechanisms must exist to establish and maintain specific enhancer–promoter interactions across large distances.
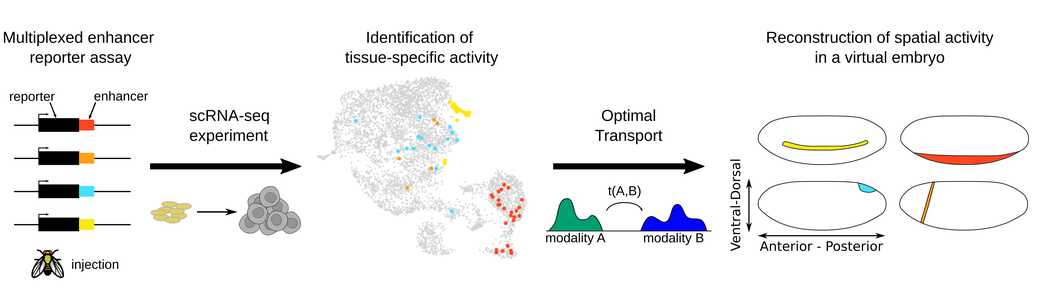
Spatial-scERA: a method for reconstructing spatial single-cell enhancer activity in multicellular organisms
Enhancers play an essential role in developmental processes by regulating the spatiotemporal expression of genes. Characterizing their spatiotemporal activity remains however an important challenge. Here we introduce a novel in vivo/in silico method for spatial single-cell enhancer-reporter assays (spatial-scERA) designed to reconstruct the spatial activity of candidate enhancer regions in parallel in multicellular organisms. Spatial-scERA integrates parallel reporter assays with single-cell RNA sequencing and spatial reconstruction using optimal transport, to map cell-type-specific enhancer activity at the single-cell level on a 3D virtual sample. We evaluated spatial-scERA in Drosophila embryos using 25 candidate enhancers, and validated the robustness of our reconstructions by comparing them to in situ hybridization. Remarkably, spatial-scERA faithfully reconstructed the spatial activity of these enhancers, even when the reporter construct was expressed in as few as 10 cells. Our results demonstrate the importance of integrating transcriptomic and spatial data for accurately predicting enhancer activity patterns in complex multicellular samples and linking enhancers to their potential target genes. Overall, spatial-scERA provides a scalable approach to map the spatial activity of enhancers at single-cell resolution without the need for imaging or a priori knowledge of embryology and can be applied to any multicellular organism amenable to transgenesis.
Experimental techniques and equipment
The main tools used in the lab are:
- genomics: 4C/Hi-C, RNA-seq, ChIP-seq, ATAC-seq, single-cell OMICs
- imaging: DNA and RNA FISH, MERFISH
- CRISPR-Cas9 genome editing
- FACS sorting
- Drosophila genetics and transgenesis
- bioinformatics
Our favorite model organisms are the fruit fly Drosophila melanogaster and the tunicate Oikopleura dioica
We benefit from the core infrastructure available at the IGFL including shared facilities for animal housing, cell culture, microscopy, an in-situ hybridization robot, and a large particle sorter (Union Biometrica BioSorter) to sort intact embryos based on size and fluorescent pattern. The IGFL has an in-house NGS service with Illumina and Nanopore technologies. Moreover, we will have access to state-of-the-art computing clusters (>500 compute nodes) and use the Core Facilities of the Gerland Campus located in close proximity to the Institute.
Funding










Institut de Génomique Fonctionnelle de Lyon (IGFL)
ENS de Lyon - CNRS UMR 5242 - INRA USC 1370
32-34 avenue Tony Garnier, 69007 Lyon, France




